Part C: Identifying Synonymous and Nonsynonymous Mutations
In order to determine if the SNP in your variant sequence will affect the structure and function of the protein, you will need to align the two amino acid sequences (like you did with the gene sequences) and determine if the SNP causes a synonymous or nonsynonymous mutation in the variant protein. Here’s how to do that.
11. Return to (or reopen) ClustalW at http://www.genome.jp/tools/clustalw/.
a. On ClustalW, next to ‘Enter your sequences…’ click on Protein.
b. Copy and paste the wild-type TB and variant TB polypeptide sequences from Word into ClustalW. Be sure each sequence is labeled with a > and a name.
c. Click ‘Execute Multiple Alignment.’
d. On the next page that comes up, scroll down to the section headed with ‘clustalw.aln.’
e. Look at the alignment of your two sequences. Stars (***) indicate amino acids that are identical. A semicolon (:), a period (.), or a blank space indicates a changed amino acid. A series of hyphens (-----) indicates missing amino acids.
12) Are your amino acid sequences identical or are they different? ____________________
Therefore, does the SNP in your variant rpoB gene sequence cause a synonymous mutation or a nonsynonymous mutation? ______________________________________
o If they are identical, skip to Question 15. If they are different, continue with Question 13.
13) For variants C-F and H, fill in the chart below to compare the different amino acid between the two sequences. For variant G, skip to Question 14.
a. Use Copymaster 1: Amino Acid Chart and Copymaster 2: Properties of Amino Acids.
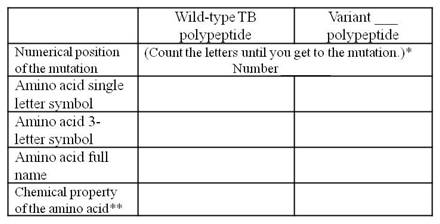
*An easy way to do this in Word is to highlight the amino acids from the beginning of the sequence, stopping when you’ve just included the different amino acid. Then, click on ‘Review’ at the top of Word, then click on ‘Word Count.’ The Word Count will give you the numerical position of the mutation.
**Click on the Properties of Amino Acids chart. Determine what chemical property
each amino acid has: acidic, basic, polar, or nonpolar.
14) For Variant G, provide and explanation for why there are missing amino acids at the end of the variant protein.
_____________________________________________________________
15) Does the amino acid change cause sense, missense or nonsense? _________________
(Refer to the Background section) Why did you classify the change that way? __________________________
Link to Part D
Copyright © 2013 Michael Strong and Jessica Taylor